New: We are pleased to announce that our work is serving as a basis for the Bacteria Gene Interactions task of the BioNLP Shared Task series. The link to the dataset below will be disabled during the competition.
You can find
here the dataset used for experiments in
the CICLing'10 paper: Extraction of Genic Interactions with the
Recursive Logical Theory of an Ontology.
Abstract:
We introduce an Information Extraction (IE) system which uses the logical theory of an ontology as a generalisation of the typical information extraction patterns to extract biological interactions from text. This provides inferences capabilities beyond current approaches: first, our system is able to handle multiple relations; second, it allows to handle dependencies between relations, by deriving new relations from the previously extracted ones, and using inference at a semantic level; third, it addresses recursive or mutually recursive rules.
In this context, automatically acquiring the resources of an IE system becomes an ontology learning task: terms, synonyms, conceptual hierarchy, relational hierarchy, and the logical theory of the ontology have to be acquired. We focus on the last point, as learning the logical theory of an ontology, and a fortiori of a recursive one, remains a seldom studied problem.
We validate our approach by using a relational learning algorithm, which handles recursion, to learn a recursive logical theory from a text corpus on the bacterium Bacillus subtilis. This theory achieves a good recall and precision for the ten defined semantic relations, reaching a global recall of 67.7% and a precision of 75.5%, but more importantly, it captures complex mutually recursive interactions which were implicitly encoded in the ontology.
The format is a collection of prolog facts. Each fact describes an instance of a relation or a concept of the domain ontology. The file is rather straightforward to read if you are familiar with prolog.
The following figure exemplifies the representation of the sentence fragment "The DNA binding protein GerE stimulates transcription [...]"
term(su162, stimulate).
term(su164, transcription).
term(su161, 'DNA binding protein GerE').
syntactic_relation(su162, su164, 'obj:V-N').
syntactic_relation(su162, su161, 'subj:V-N').
lexical_ref('migal85', su162).
lexical_ref('migal89', su164).
lexical_ref('migal90', su161).
instance('migal85', reg).
instance('migal89', tra).
instance('migal90', p).
semantic_relation('migal90', 'migal89', t_by).
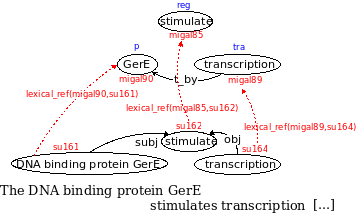 1) Syntactic level:
The terms of the sentence are identified and assigned to an unique ID (i.e. "su161" for the term "DNA binding protein GerE"):
term(su162, stimulate).
term(su164, transcription).
term(su161, 'DNA binding protein GerE').
Syntactic relations link terms between them (i.e. an ('obj' (object) relation between su162 and su164):
syntactic_relation(su162, su164, 'obj:V-N').
syntactic_relation(su162, su161, 'subj:V-N').
(Note: The semantic of labels is the same as for the LLL'05 Challenge.)
2) Lexical Layer:
Each term is linked to the semantic level through the "lexical_ref" relation, which associates a term ID to an ID of a concept's instance.
lexical_ref('migal85', su162).
lexical_ref('migal89', su164).
lexical_ref('migal90', su161).
3) Conceptual level:
Each instance has a unique ID: for example, "stimulate" becomes "migal85", an instance of the concept "reg" (regulation).
instance('migal85', reg).
instance('migal89', tra).
instance('migal90', p).
Instances of concept are linked by semantic relations (i.e. "migal90" and "migal89" by a "t_by" ("transcription by" relation)).
semantic_relation('migal90', 'migal89', t_by).
1) Syntactic level:
The terms of the sentence are identified and assigned to an unique ID (i.e. "su161" for the term "DNA binding protein GerE"):
term(su162, stimulate).
term(su164, transcription).
term(su161, 'DNA binding protein GerE').
Syntactic relations link terms between them (i.e. an ('obj' (object) relation between su162 and su164):
syntactic_relation(su162, su164, 'obj:V-N').
syntactic_relation(su162, su161, 'subj:V-N').
(Note: The semantic of labels is the same as for the LLL'05 Challenge.)
2) Lexical Layer:
Each term is linked to the semantic level through the "lexical_ref" relation, which associates a term ID to an ID of a concept's instance.
lexical_ref('migal85', su162).
lexical_ref('migal89', su164).
lexical_ref('migal90', su161).
3) Conceptual level:
Each instance has a unique ID: for example, "stimulate" becomes "migal85", an instance of the concept "reg" (regulation).
instance('migal85', reg).
instance('migal89', tra).
instance('migal90', p).
Instances of concept are linked by semantic relations (i.e. "migal90" and "migal89" by a "t_by" ("transcription by" relation)).
semantic_relation('migal90', 'migal89', t_by).
 1) Syntactic level:
The terms of the sentence are identified and assigned to an unique ID (i.e. "su161" for the term "DNA binding protein GerE"):
term(su162, stimulate).
term(su164, transcription).
term(su161, 'DNA binding protein GerE').
Syntactic relations link terms between them (i.e. an ('obj' (object) relation between su162 and su164):
syntactic_relation(su162, su164, 'obj:V-N').
syntactic_relation(su162, su161, 'subj:V-N').
(Note: The semantic of labels is the same as for the LLL'05 Challenge.)
2) Lexical Layer:
Each term is linked to the semantic level through the "lexical_ref" relation, which associates a term ID to an ID of a concept's instance.
lexical_ref('migal85', su162).
lexical_ref('migal89', su164).
lexical_ref('migal90', su161).
3) Conceptual level:
Each instance has a unique ID: for example, "stimulate" becomes "migal85", an instance of the concept "reg" (regulation).
instance('migal85', reg).
instance('migal89', tra).
instance('migal90', p).
Instances of concept are linked by semantic relations (i.e. "migal90" and "migal89" by a "t_by" ("transcription by" relation)).
semantic_relation('migal90', 'migal89', t_by).
1) Syntactic level:
The terms of the sentence are identified and assigned to an unique ID (i.e. "su161" for the term "DNA binding protein GerE"):
term(su162, stimulate).
term(su164, transcription).
term(su161, 'DNA binding protein GerE').
Syntactic relations link terms between them (i.e. an ('obj' (object) relation between su162 and su164):
syntactic_relation(su162, su164, 'obj:V-N').
syntactic_relation(su162, su161, 'subj:V-N').
(Note: The semantic of labels is the same as for the LLL'05 Challenge.)
2) Lexical Layer:
Each term is linked to the semantic level through the "lexical_ref" relation, which associates a term ID to an ID of a concept's instance.
lexical_ref('migal85', su162).
lexical_ref('migal89', su164).
lexical_ref('migal90', su161).
3) Conceptual level:
Each instance has a unique ID: for example, "stimulate" becomes "migal85", an instance of the concept "reg" (regulation).
instance('migal85', reg).
instance('migal89', tra).
instance('migal90', p).
Instances of concept are linked by semantic relations (i.e. "migal90" and "migal89" by a "t_by" ("transcription by" relation)).
semantic_relation('migal90', 'migal89', t_by).